Colibri Core Documentation¶
Introduction¶
Colibri Core is software consisting of command line tools as well as programming libraries. to quickly and efficiently count and extract patterns from large corpus data, to extract various statistics on the extracted patterns, and to compute relations between the extracted patterns. The employed notion of pattern or construction encompasses the following categories:
n-gram – n consecutive words
skipgram – An abstract pattern of predetermined length with one or multiple gaps (of specific size).
flexgram – An abstract pattern without predetermined length, with one or more gaps.
N-gram extraction may seem fairly trivial at first, with a few lines in your favourite scripting language, you can move a simple sliding window of size n over your corpus and store the results in some kind of hashmap. This trivial approach however makes an unnecessarily high demand on memory resources, this often becomes prohibitive if unleashed on large corpora. Colibri Core tries to minimise these space requirements in several ways:
Binary representation – Each word type is assigned a numeric class, which is encoded in a compact binary format in which highly frequent classes take less space than less frequent classes. Colibri core always uses this representation rather than a full string representation, both on disk and in memory.
Informed counting – Counting is performed more intelligently by iteratively processing the corpus in several passes and quickly discarding patterns that won’t reach the desired occurrence threshold.
Skipgram and flexgram extraction are computationally more demanding but have been implemented with similar optimisations. Skipgrams are computed by abstracting over n-grams, and flexgrams in turn are computed either by abstracting over skipgrams, or directly from n-grams on the basis of co-occurrence information (mutual pointwise information). When patterns have been extracted, along with their counts and or index references to original corpus data, they form a so-called pattern model.
At the heart of Colibri Core lies the tool colibri-patternmodeller which
allows you to build, view, manipulate and query pattern models.
The Colibri software is developed in the scope of the Ph.D. research project Constructions as Linguistic Bridges. This research examines the identification and extraction of aligned constructions or patterns across natural languages, and the usage of such constructions in Machine Translation. The aligned constructions are not identified on the basis of an extensive and explicitly defined grammar or expert database of linguistic knowledge, but rather are implicitly distilled from large amounts of example data. Our notion of constructions is broad and transcends the idea of words or variable-length phrases.
This documentation will illustrate how to work with the various tools and the library of colibri, as well as elaborate on the implementation of certain key aspects of the software.
Installation¶
Installing dependencies¶
To compile Colibri Core, you need a sane build environment, install the necessary dependencies for your distribution.
For Debian/Ubuntu:
$ sudo apt-get install make gcc g++ pkg-config autoconf-archive libtool autotools-dev libbz2-dev zlib1g-dev libtar-dev python3 python3-dev cython3"
For RedHat-based systems (run as root):
# yum install pkgconfig libtool autoconf automake autoconf-archive make gcc gcc-c++ libtar libtar-devel python3 python3-devel zlib zlib-devel python3-pip bzip2 bzip2-devel cython3
For Mac OS X with homebrew:
$ brew install autoconf automake libtool autoconf-archive python3 pkg-config
Arch Linux users can simply install Colibri Core and all dependencies directly from the Arch User Repository , no further installation is necessary in this case.
Installation via the Python Package Index¶
Colibri Core can be installed from the Python Package Index using the pip tool (may be named pip3).
This procedure will automatically download, compile, and install all of Colibi Core.
First ensure you installed all dependencies from the first section!
Colibri Core requires an up-to-date version of Cython first (0.23 or above), or the installation will fail, we use pip to compile it from scratch:
$ sudo pip3 install cython
Then we can install Colibri Core itself:
$ sudo pip3 install colibricore
For installation without root privileges we recommend creating a Python Virtual environment, , in which case all of Colibri Code will be installed under it:
$ python3 -m venv env
$ . env/bin/activate #you will need to do this each time you want to use Colibri Core
(env)$ pip install cython
(env)$ pip install colibricore
Important Note: If you install Colibri Core locally (in a Python Virtual Environment
or elsewhere), then you need to set LD_LIBRARY_PATH=$VIRTUAL_ENV/lib/ prior
to running Python for the Python binding to function (replace $VIRTUAL_ENV with
the directory you used as a prefix if you do not use Python Virtual
Environment). Otherwise, you will be confronted with an error: ImportError:
libcolibricore.so.0: cannot open shared object file: No such file or
directory. If you get this error after a global installaton, run sudo
ldconfig.
Installation from Github¶
First ensure you installed all dependencies from the dependencies section!
Colibri Core is hosted on github
and should ideally be retrieved through the versioning control system git.
Provided git is installed on your system, this is done as follows:
$ git clone https://github.com/proycon/colibri-core.git
$ cd colibri-core
Alternatively, you can download and extract release archives from the aforementioned Github page.
If you install from within a Python Virtual environment, everything will be installed under it. This does not require root privileges:
$ python3 -m venv env
$ . env/bin/activate #you will need to do this each time you want to use Colibri Core
(env)$ pip install cython
(env)$ pip install .
The note at the end of the previous section applies for any non-global installation!
Container image¶
If you are interested in only the command-line tools (not the Python binding) and you don’t want to compile from source, then an OCI container image is available for use with for example Docker:
$ docker pull proycon/colibri-core
Manual compilation and installation (advanced)¶
Installation via the Python Package Index or Github invokes compilation of the C++ source for you. You can however to compile everything yourself. This is especially relevant if you are not only interested in command-line tools or C++ library, and do not care about the Python library.
Moreover, this route is also needed if you want FoLiA support (optional), for which you then first to install the following dependency. By default FoLiA support is disabled.
libfolia; obtainable from the FoLiA website, follow the instructions included with libfolia to install it.
Including pulling the sources from github, Colibri Core can be compiled and installed as follows:
$ git clone https://github.com/proycon/colibri-core.git
$ cd colibri-core
$ bash bootstrap
$ ./configure [--prefix=/usr] [--with-folia --with-folia-path=/path/to/libfolia]
$ make
$ sudo make install
If not prefix is set, installation will be under /usr/local/ by default.
To compile the Python binding manually, we first create an empty file
manual which signals the build process to not attempt to recompile the C++
library itself:
$ touch manual
Then build as follows, provided Colibri Core is in a globally accessible location already:
$ sudo python3 ./setup.py install
If you used a prefix and want to install in a customised non-global location,
again use --prefix along with --include-dirs and --library-dirs to
point to where the C++ headers and the library are installed:
$ python3 ./setup.py build_ext --include-dirs=/path/to/include/colibri-core --library-dirs=/path/to/lib/ install --prefix=/path/to/somewhere/
Update $LD_LIBRARY_PATH and $PYTHONPATH where necessary.
Keeping colibri up to date¶
It is recommended to often check back for new versions of Colibri Core.
If you used, pip just run:
$ pip3 install -U colibricore
For the git version:
$ git pull
For this one you’ll need to recompile as per the above instructions.
For the docker image:
$ docker pull proycon/colibricore
General usage instructions¶
Colibri consist of various programs and scripts, each of which will output an
extensive overview of available parameters if the parameter -h is passed.
Each program is designed for a specialised purpose, with specific input and
output formats. It is often needed to call multiple programs in succession to
obtain the final analysis or model you desire.
Quick start: High-level scripts¶
Introduction¶
Colibri Core comes with a set of scripts that provide simpler access to the underlying tools and can be used from the command line by end-users to get quick results. Input to these tools is always one or more plain text files, in tokenised form, with one sentence per line.
Tokenisation¶
If your corpus is not tokenised yet, you can consider using the tokeniser ucto , which is not part of Colibri Core, Debian/Ubuntu
users may it in the repository (sudo apt-get install ucto), Mac OS X users
can find it in homebrew (brew install naiaden/lama/ucto). This will also do
sentence detection and, with the -n flag output one line per sentence, as
Colibri prefers:
$ ucto -L en -n untokenisedcorpus.txt > tokenisedcorpus.txt
The -L specifies the language of your corpus (English in this case), several others are
available as well.
Of course, you can use any other tokeniser of your choice.
Scripts¶
In addition to the core tools, described in the remainder of this documentation, Colibri Core offers the following scripts:
colibri-ngrams- Extracts n-grams of a particular size from the corpus text, in the order they occur, i.e. by moving a sliding window over the text.
colibri-freqlist- Extracts all n-grams from one or more corpus text files andoutputs a frequency list. Also allows for the extraction of skipgrams. By default all n-grams are extracted, but an occurrence threshold can be set with the
-tflag.
colibri-ngramstats- Prints a summary report on the ngrams in one ormore corpus text files. To get the full details on interpreting the output report, read the section Statistical Reports and Histograms.
colibri-histogram- Prints a histogram of ngram/skipgram occurrence count
colibri-queryngrams- Interactive tool allowing you to query ngrams from standard input, various statistics and relations can be output.
colibri-reverseindex- Computes and prints a reverse index for the specified corpus text file. For each token position in the corpus, it will output what patterns are found there (i.e start at that very same position)
colibri-loglikelihood- Computes the log-likelihood between patterns in two or more corpus text files, which allows users to determine what words or patterns are significantly more frequent in one corpus than the other.
colibri-coverage- Computes overlap between a training corpus and a test corpus, produces coverage metrics.
colibri-findpatterns- Finds patterns (including skipgrams and flexgrams) in corpus data. You specify the patterns in a text file (one per line).
Users have to be aware, however, that these script only expose a limited amount of the functionality of Colibri Core. Nevertheless, they simplify a lot of common tasks people do with Colibri Core.
Corpus Class Encoding¶
Introduction¶
Computation on large datasets begs for solutions to keep memory consumption manageable. Colibri requires that input corpora are converted into a compressed binary form. The vocabulary of the corpus is converted to integer form, i.e. each word-type in the corpus is represented by a numeric class. Highly frequent word-types get assigned low class numbers and less frequent word-types get higher class numbers. The class is represented in a dynamic-width byte-array, rather than a fixed-width integer. Patterns are encoded per word, each word starts with a size marker of one byte indicating the number of bytes are used for that word. The specified number of bytes that follow encode the word class. Instead of a size marker, byte values of 128 and above are reserved for special markers, such as encoding gaps and structural data. Finally, the pattern as a whole is ended by a null byte.
All internal computations of all tools in colibri proceed on this internal representation rather than actual textual strings, keeping running time shorter and memory footprint significantly smaller.
Class-encoding your corpus¶
When working with colibri, you first want to class encode your corpus. This is done by the program colibri-classencode. It takes as input a tokenised monolingual corpus in plain text format, containing one sentence per line, as a line is the only structural unit Colibri works with, extracted patterns will never cross line boundaries. Each line should be delimited by a single newline character (unix line endings). If you desire another structural unit (such as for example a tweet, or a paragraph), simply make sure each is on one line.
Colibri is completely agnostic when it comes to the character encoding of the input. Given a corpus file yourcorpus, class encoding is done as follows:
$ colibri-classencode yourcorpus
This results in two files:
yourcorpus.colibri.cls- This is the class file; it lists all word-types and class numbers.
yourcorpus.colibri.dat- This is the corpus is encoded binary form. It is a lossless compression that is roughly half the size of the original
If your corpus is not tokenised yet, you can consider using the tokeniser ucto (not part of colibri), this will also do sentence detection and output one line per sentence:
$ ucto -L en -n untokenisedcorpus.txt > tokenisedcorpus.txt
The above sample is for English (-L en), several other languages are also supported.
In addition to this plain text input. The class encoder also supports FoLiA
XML (folia website) if you compiled with
FoLiA support, make sure such files end with the extension xml and they
will be automatically interpreted as FoLiA XML:
$ colibri-classencode yourcorpus.xml
The class file is the vocabulary of your corpus, it simply maps word strings to integer. You must always ensure that whenever you are working with multiple models, and you want to compare them, to use the exact same class file. It is possible to encode multiple corpus files similtaneously, generating a joined class file:
$ colibri-classencode yourcorpus1.txt yourcorpus2.txt
This results in yourcorpus1.colibri.cls and yourcorpus1.colibri.dat and
yourcorpus2.colibri.dat. The class file spans both despite the name. An
explicit name can be passed with the -o flag. It is also possible to encode
multiple corpora in a single unified file by passing the -u flag. This is
often desired if you want to train a pattern model on all the joined data:
$ colibri-classencode -o out -u yourcorpus1.txt yourcorpus2.txt
This will produce out.colibri.dat and out.colibri.cls. You can use the -l option to read input filenames from file instead of command line arguments (one filename per line).
If you have a pre-existing class file you can load it with the -c flag, and use it to encode new data:
$ colibri-classencode -c yourcorpus1.colibri.cls yourcorpus2.txt
This will produce a yourcorpus2.colibri.dat, provided that all of the word
types already existed in yourcorpus1.colibri.cls (which usually is not the
case, in which case an error will be shown. To circumvent this error you have
to specify how to deal with unknown words. There are two ways; the -U flag
will encode all unknown word as a single word class dedicated to the task,
whereas the -e flag will extend the specified class file with any new
classes found. It has to be noted that this extension method spoils the optimal
compression as classes are no longer strictly sorted by frequency. If you can
all needed data in one go, then that is always preferred.
This setup, however, is often seen in a train/test paradigm:
$ colibri-classencode -f testset.txt -c trainset.colibri.cls -e
This will result in an encoded corpus testset.colibri.dat and an extended class file testset.colibri.cls, which is a superset of the original trainset.cls, adding only those classes that did not yet exist in the training data.
Class-decoding your corpus¶
Given an encoded corpus and a class file, the original corpus can always be reconstructed (unless the -U option was used in encoding to allow unknown words). This we call class decoding and is done using the colibri-classdecode program:
$ colibri-classdecode -f yourcorpus.colibri.dat -c yourcorpus.colibri.cls
Partial decoding can be done by specifying start and end line numbers using the
flags -s and -e respectively.
Output will be to stdout, you can redirect it to a file as follows:
$ colibri-classdecode -f yourcorpus.colibri.dat -c yourcorpus.colibri.cls > yourcorpus.txt
Pattern Modeller¶
Introduction¶
The colibri-patternmodeller program is used to create pattern models
capturing recurring patterns from a monolingual corpus. The extracted patterns
are n-grams or skip-grams, where a skip-gram is an n-gram with one or more gaps
of either a predefined size, thus containing unspecified or wildcard tokens, or
of dynamic width.
In the internal pattern representation, in the place of the size marker, byte value 128 is used for a fixed gap of a single token, and can be repeated for gaps of longer length, byte value 129 is used for a gap of unspecified dynamic width.
The pattern finding algorithm is iterative in nature and is guaranteed to find all n-grams above a specified occurrence threshold and optionally given a maximum size for n. It does so by iterating over the corpus n times, iterating over all possible values for n in ascending order. At each iteration, a sliding window extracts all n-grams in the corpus for the size is question. An n-gram is counted in a hashmap data structure only if both n-1-grams it by definition contains are found during the previous iteration with an occurrence above the set threshold. The exception are unigrams, which are all by definition counted if they reach the threshold, as they are already atomic in nature. At the end of each iteration, n-grams not making the occurrence threshold are pruned. This simple iterative technique reduces the memory footprint compared to the more naive approach of immediately storing all in a hashmap, as it prevents the storing of lots of patterns not making the threshold by discarding them at an earlier stage.
At the beginning of each iteration of n, all possible ways in which any n-gram of size n can contain gaps is computed. When an n-gram is found, various skip-grams are tried in accordance with these gap configurations. This is accomplished by ‘punching holes’ in the n-gram, resulting in a skip-gram. If all consecutive parts of this skip-gram were counted during previous iterations and thus made the threshold, then the skip-gram as a whole is counted, otherwise it is discarded. After each iteration, pruning again takes places to prune skip-grams that are not frequent enough.
The pattern finder can create either indexed or unindexed models. For indexed models, the precise location of where an n-gram or skipgram instance was found in the corpus is recorded. This comes at the cost of much higher memory usage, but is necessary for more strongly constrained skip extraction, as well as for extracting relations between patterns at a later stage. Indexed models by default also maintain a reverse index allowing, and even unindexed models do so during building.
Note that for fixed-size skipgrams in indexed models, the various fillings for the gaps can be reconstructed precisely.
If you are only interested in simple n-gram or simple skip-gram counts, then an unindexed model may suffice.
Creating a pattern model¶
First make sure to have class-encoded your corpus. Given this encoded corpus,
colibri-patternmodeller can be invoked to produce an indexed pattern model.
Always specify the output file using the --outputmodel or -o flag. The occurrence threshold
is specified with parameter --threshold or -t, patterns occuring less will not be counted.
The default value is two. The maximum value for n, i.e. the maximum
n-gram/skipgram size, can be restricted using the parameter -l.:
$ colibri-patternmodeller --datafile yourcorpus.dat --threshold 10 --outputmodel yourcorpus.colibri.indexedpatternmodel
This outputted model yourcorpus.colibri.indexedpatternmodel is stored in a
binary format. To print it into a human readable presentation it needs to be
decoded. The colibri-patternmodeller program can do this by specifying an
input model using the --inputmodel or -i flag, the class file using the --classfile (-c) parameter,
and the desired action to print it all using --print (-P):
$ colibri-patternmodeller --inputmodel yourcorpus.colibri.indexedpatternmodel --classfile yourcorpus.colibri.cls --print
Optionally, instead of or in addition to outputting a model to file using
-o, you can also print it directly with -P.
Output will be to stdout in a tab delimited format, with the first line
reserved for the header. This facilitates easy parsing as you can just load it
into any software accepting CSV files, such as spreadsheets. An excerpt
follows:
PATTERN COUNT TOKENS COVERAGE CATEGORY SIZE FREQUENCY REFERENCES
For 2 2 0.0059 ngram 1 0.0121 11:0 15:0
death 2 2 0.0059 ngram 1 0.0121 11:5 23:7
who 2 2 0.0059 ngram 1 0.0121 15:1 21:5
. 4 4 0.0118 ngram 1 0.0242 5:6 9:4 10:6 13:4
be 4 4 0.0118 ngram 1 0.0242 1:1 1:5 9:2 35:3
flee 2 2 0.0059 ngram 1 0.0121 36:1 36:5
not to 4 8 0.0235 ngram 2 0.1538 1:3 36:3 37:3 38:3
The various columns are:
Pattern - The actual pattern. Gaps in skipgrams are represented as
{*}(for each gap). Variable-width skipgrams are just{**}.Occurrence count - The absolute number of times this pattern occurs
Tokens - The absolute number of tokens in the corpus that this pattern covers. Longer patterns by definition cover more tokens. This value’s maximum is
occurrencecount * n, the value will be smaller if a pattern overlaps itself.Coverage - The number of covered tokens, as a fraction of the total number of tokens.
Category - The type of pattern (ngram, skipgram or flexgram).
Size - The length of the n-gram or skipgram in words/tokens.
Frequency - The frequency of the pattern within its category and size class, so for an ngram of size two, the frequency indicates the frequency amongst all bigrams.
References - A space-delimited list of indices in the corpus that correspond to a occurrence of this pattern. Indices are in the form
sentence:tokenwhere sentence starts at one and token starts at zero. This column is only available for indexed models.
Creating a pattern model with skipgrams and/or flexgrams¶
The pattern model created in the previous example did not yet include
skip-grams, these have to be explicitly enabled with the --skipgrams (-s) flag. When this
is used, another options becomes available for consideration:
--skiptypes [value](-Tfor short) - Only skipgrams that have at least this many different types as skip content, i.e. possible options filling the gaps, will be considered. The default is set to two.
Here is an example of generating an indexed pattern model including skipgrams:
$ colibri-patternmodeller --datafile yourcorpus.colibri.dat --threshold 10 --skipgrams --skiptypes 3 --outputmodel yourcorpus.colibri.indexedpatternmodel
If you want to generate unindexed models, simply add the flag --unindexed or -u for short. Do note
that for unindexed models the parameter --skiptypes has no effect, it will extract
all skipgrams it can find as if -skiptypes were set to one! If you want decent
skipgrams, you’re best off with an indexed model. Note that indexed models can
always be read and printed in an unindexed way (with the --unindexed flag); but
unindexed models can not be read in an indexed way, as they simply lack
indices:
$ colibri-patternmodeller --inputmodel yourcorpus.colibri.indexedpatternmodel --classfile yourcorpus.colibri.cls --unindexed --print
$ colibri-patternmodeller --inputmodel yourcorpus.colibri.unindexedpatternmodel --classfile yourcorpus.colibri.cls --unindexed --print
Flexgrams, non-consecutive patterns in which the gap (only one in the current implementation) is of dynamic width, can be generated in one of two ways:
Extract flexgrams by abstracting from skipgrams: use the
--flexgrams Sflag.Extract flexgrams directly from n-gram co-occurence: use the
--flexgams [threshold]flag, where the threshold is expressed as normalised pointwise mutual information [-1,1].
The skipgram approach has the advantage of allowing you to rely on the
--skiptypes threshold, but comes with the disadvantage of having a maximum span. The
co-occurrence approach allows for flexgrams over larger distances. Both methods
come at the cost of more memory, especially the former method.
Neither skipgrams nor flexgrams will cross the line boundary of the original corpus data, so ensure your data is segmented into lines suitable for your purposes in the encoding stage.
Two-stage building¶
Generating an indexed pattern model takes considerably more memory than an unindexed model, as instead of mere counts, all indices have to be retained.
The creation of a pattern models progresses through stages of counting and pruning. When construction of an indexed model with an occurrence threshold of 2 or higher reaches the limits of your system’s memory capacity, then two-stage building may offer a solution.
Two-stage building first constructs an unindexed model (demanding less memory),
and subsequently loads this model and searches the corpus for indices for all
patterns in the model. Whilst this method is more time-consuming, it prevents
the memory bump (after counting, prior to pruning) that normal one-stage
building of indexed models have. Two-stage building is enabled using the -2 flag:
$ colibri-patternmodeller -2 --datafile yourcorpus.colibri.dat --threshold 10 --skipgrams --skiptypes 3 -outputmodel yourcorpus.colibri.indexedpatternmodel
Statistical reports and histograms¶
If you have a pattern model, you can generate a statistical report which includes information on the number of occurrences and number of types for patterns, grouped for n-grams or skipgrams for a specific value of n. A report is generated using the -report (-R) flag, the input model is specified using --inputmodel (-i):
$ colibri-patternmodeller --inputmodel yourcorpus.colibri.indexedpatternmodel --report
Example output:
REPORT
----------------------------------
PATTERNS TOKENS COVERAGE TYPES
Total: - 340 - 177
Uncovered: - 175 0.5147 136
Covered: 69 165 0.4853 41
CATEGORY N (SIZE) PATTERNS TOKENS COVERAGE TYPES OCCURRENCES
all all 69 165 0.4853 41 243
all 1 40 165 0.4853 40 165
all 2 11 26 0.0765 13 26
all 3 7 17 0.0500 9 19
all 4 5 10 0.0294 9 14
all 5 5 9 0.0265 9 17
all 6 1 2 0.0059 6 2
n-gram all 62 165 0.4853 40 215
n-gram 1 40 165 0.4853 40 165
n-gram 2 11 26 0.0765 13 26
n-gram 3 5 12 0.0353 8 12
n-gram 4 3 6 0.0176 6 6
n-gram 5 2 4 0.0118 6 4
n-gram 6 1 2 0.0059 6 2
skipgram all 7 7 0.0206 6 28
skipgram 3 2 7 0.0206 4 7
skipgram 4 2 4 0.0118 4 8
skipgram 5 3 5 0.0147 5 13
Some explanation is in order to correctly interpret this data. First of all patterns are grouped by category (ngram,skipgram, flexgram) and size. There are various metrics:
Pattern - The number of distinct patterns in this group, so for category n-gram of 2, this reflects the number of distinct bigrams.
Tokens - The number of tokens that is covered by the patterns in the group. Longer patterns by definition cover more tokens. . This is only available for indexed models, for unindexed models it is either omitted or the number shown is maximum projection
occurrencecount * size.Coverage - The number of tokens covered as a fraction of the total number of tokens. Only for indexed models.
Types - The number of unique word types covered, i.e the number of distinct unigrams.
Occurrences - Cumulative occurrence count of all the patterns in the group. Used as a basis for computing frequency. Occurrence count differs from tokens, the former expresses the number of times a pattern occurs in the corpus, the latter expresses how many tokens are part of the pattern
To better understand these metrics, let’s perceive them in the following test sentence:
to be or not to be , that is the question
If we generate an indexed pattern model purely on this sentence, with threshold two. We find the following three patterns:
PATTERN COUNT TOKENS COVERAGE CATEGORY SIZE FREQUENCY REFERENCES
to 2 2 0.181818 ngram 1 0.5 1:0 1:4
be 2 2 0.181818 ngram 1 0.5 1:1 1:5
to be 2 4 0.363636 ngram 2 1 1:0 1:4
The report then looks as follows:
REPORT
----------------------------------
PATTERNS TOKENS COVERAGE TYPES
Total: - 11 - 9
Uncovered: - 7 0.6364 7
Covered: 3 4 0.3636 2
CATEGORY N (SIZE) PATTERNS TOKENS COVERAGE TYPES OCCURRENCES
all all 3 4 0.3636 2 6
all 1 2 4 0.3636 2 4
all 2 1 4 0.3636 2 2
n-gram all 3 4 0.3636 2 6
n-gram 1 2 4 0.3636 2 4
n-gram 2 1 4 0.3636 2 2
Our sentence has 11 tokens, 7 of which are not covered by the patterns found, 4 of which are. Since we have only n-grams and no skipgrams or flexgrams in this simple example, the data for all and n-gram is the same. The coverage metric expresses this in a normalised fashion.
In our data we have two unigrams (to, be) and one bigram (to be), this is expressed by the patterns metric. Both the unigrams and the bigrams cover the exact same four tokens in our sentence, i.e 0, 1, 4, and 5, so the TOKENS column reports four for all. If we look at the types column, we notice we only have two word types: to and be. The unigrams occur in four different instances and the bigrams occur in two different instances. This is expressed in the occurrences column. Combined that makes six occurrences.
We have 9 types in total, of which only 2 (to, be) are covered, the remaining 7 (or not , that is the question) remain uncovered as we set our occurrence threshold for this model to two.
Pattern models store how many of the tokens and types in the original corpus were covered. Tokens and types not covered did not make the set thresholds. Make sure to use indexed models if you want accurate coverage data.
A histogram can also be generated, using the --histogram (-H) flag:
$ colibri-patternmodeller --inputmodel yourcorpus.colibri.indexedpatternmodel --histogram
Example output:
OCCURRENCES PATTERNS
2 39
3 5
4 13
5 5
6 1
7 1
8 1
10 1
13 1
14 1
15 1
Filtering models¶
Patterns models can be read with --inputmodel and filtered by setting stricter thresholds prior to printing, reporting or outputting to file. An example:
$ colibri-patternmodeller --inputmodel yourcorpus.colibri.indexedpatternmodel -t 20 --skiptypes 10 --outputmodel yourcorpus_filtered.colibri.indexedpatternmodel --print
You can also filter pattern models by intersecting with another pattern model
using the --constraintmodel option. This only works when both are built on
the same class file:
$ colibri-patternmodeller --inputmodel yourcorpus.colibri.indexedpatternmodel --constraintmodel yourcorpus2.colibri.indexedpatternmodel --outputmodel yourcorpus_filtered.colibri.indexedpatternmodel
The output pattern model will contain only those patterns that were present in
both the input model (--inputmodel) as well as the constraining model
(--constraintmodel), which may be either indexed or unindexed regardless of
the input model; it will always contain the counts/indices from the input
model.
Training and testing coverage¶
An important quality of pattern models lies in the fact that pattern models can
be compared, provided they use comparable vocabulary, i.e. are based on the
same class file. More specifically, you can train a pattern model on a corpus
and test it on another corpus, which yields another pattern model containing
only those patterns that occur in both training and test data. The difference
in count, frequency and coverage can then be easily be compared. You build such
a model by taking the intersection with a training model using the
--constraintmodel flag. Make sure to always use the same class file for
all datasets you are comparing. Instructions for this were given in
classencodetraintest.
- Training::
$ colibri-patternmodeller –datafile trainset.colibri.dat –outputmodel trainset.colibri.indexedpatternmodel
This results in a model trainset.colibri.indexedpatternmodel. Now proceed
with testing on another corpus:
- Testing::
$ colibri-patternmodeller –datafile testset.colibri.dat –constraintmodel trainset.indexedpatternmodel.colibri –outputmodel testset.colibri.indexedpatternmodel
It is, however, more memory efficient to load the constraint model as the inputmodel (using --inputmodel), and specify --constrained, this will do an in-place rebuilding of the model:
$ colibri-patternmodeller --datafile testset.colibri.dat --constrained trainset.indexedpatternmodel.colibri --outputmodel testset.colibri.indexedpatternmodel
This results in a model testset.colibri.indexedpatternmodel that only contains patterns that also occur in the specified training model.
Such an intersection of models can also be created at any later stage using
--inputmodel and --constraintmodel, as shown in the previous section.
If you are interested in coverage metrics for a training model on a test model,
the colibri-coverage script provides a shortcut for this.
Reverse index¶
Indexed pattern models have a what is called a forward index. For each
pattern, all of the positions, (sentence,token), at which a token of the
pattern can be found, is held. In Colibri-core, sentences always start at 1, whereas
tokens start at 0.
A reverse index is a mapping of references of the type (sentence,token) to a set of all the
patterns that begin at that location. Such a reverse index can be constructed
from the forward index of an indexed pattern model, or it can be explicitly
given by simply passing the original corpus data to the model, which makes
reverse indices available even for unindexed models. Explicitly providing a
reverse index makes loading a model faster (especially on larger models), but at
the cost of higher memory usage, especially in case of sparse models. The usual
--datafile flag takes enables a reverse index by definition.
To compute and display the full reverse index, use the --printreverseindex (-Z) flag:
$ colibri-patternmodeller --inputmodel yourcorpus.indexedpatternmodel.colibri --datafile yourcorpus.colibri.dat --classfile yourcorpus.colibri.cls --printreverseindex
Indexes and/or reverse indexes are required for various purposes, one of which is the extraction of relations and co-occurrence information.
The colibri-reverseindex script provides a shortcut for this.
Query mode¶
The pattern modeller has query mode which allows you to quickly extract
patterns from test sentences or fragments thereof. The query mode is invoked by
loading a pattern model (--inputmodel), a class file (--classfile) and
the --querymode (-Q) flag. The query mode can be run interactively as
it takes input from stdin, one tokenised sentence per line. The following
example illustrates this, the sentence “To be or not to be” was typed as
input:
$ colibri-patternmodeller --inputmodel /tmp/data.colibri.patternmodel --classfile /tmp/hamlet.colibri.cls --querymode
Loading class decoder from file /tmp/hamlet.colibri.cls
Loading class encoder from file /tmp/hamlet.colibri.cls
Loading indexed pattern model /tmp/data.colibri.patternmodel as input model...
Colibri Patternmodeller -- Interactive query mode.
Type ctrl-D to quit, type X to switch between exact mode and extensive mode (default: extensive mode).
1>> To be or not to be
1:0 To 8 8 0.0235294 ngram 1 0.0484848 1:0 5:7 9:5 10:0 22:0 36:0 37:0 38:0
1:1 be 4 4 0.0117647 ngram 1 0.0242424 1:1 1:5 9:2 35:3
1:2 or 4 4 0.0117647 ngram 1 0.0242424 1:2 36:2 37:2 38:2
1:3 not 5 5 0.0147059 ngram 1 0.030303 1:3 27:7 36:3 37:3 38:3
1:4 to 13 13 0.0382353 ngram 1 0.0787879 1:4 2:6 4:1 5:10 6:7 8:4 9:1 9:8 10:4 27:2 36:4 37:4 38:4
1:2 or not 4 8 0.0235294 ngram 2 0.153846 1:2 36:2 37:2 38:2
1:3 not to 4 8 0.0235294 ngram 2 0.153846 1:3 36:3 37:3 38:3
1:2 or not to 4 12 0.0352941 ngram 3 0.333333 1:2 36:2 37:2 38:2
The output starts with an index in the format sentence:token, specifying
where the pattern found was found in your input. The next columns are the same
as the print output.The interactive query mode distinguishes two modes,
extensive mode and exact mode. In extensive mode, your input string will be
scanned for all patterns occurring in it. In exact mode, the input you
specified needs to match exactly and as a whole. Type X to switch between
the modes.
In addition to interactive query mode, there is also a command line query mode
--query (-q) in which you specify the pattern you want to query as argument on the command
line. Multiple patterns can be specified by repeating the -q flag. This
mode always behaves according to exact mode:
$ colibri-patternmodeller --inputmodel /tmp/data.colibri.patternmodel --classfile /tmp/hamlet.colibri.cls --query "to be"
Loading class decoder from file /tmp/hamlet.colibri.cls
Loading class encoder from file /tmp/hamlet.colibri.cls
Loading indexed pattern model /tmp/data.colibri.patternmodel as input model...
to be 2 4 0.0117647 ngram 2 0.0769231 1:4 9:1
If you are working with skipgrams or flexgrams and want to explicitly
instantiate them to ngrams in the output, add the --instantiate flag.
Pattern Relations¶
A pattern model contains a wide variety of patterns; the relationships between those can be made explicit. These relationships can be imagined as a directed graph, in which the nodes represent the various patterns (n-grams and skipgrams), and the edges represent the relations. The following relations are distinguished; note that as the graph is directed relations often come in pairs; one relationship for each direction:
Subsumption relations - Patterns that are subsumed by larger patterns are called subsumption children, the larger patterns are called subsumption parents. These are the two subsumption relations that can be extracted from an indexed pattern model.
Successor relations - Patterns that follow eachother are in a left-of/right-of relation.
Instantiation relations - There is a relation between skipgrams and patterns that instantiate them
to be {*1*} not {*1*} beis instantiated byto {*1*} or, also referred to as the skip content. Similarly,to be or not to beis a full instantation, and the skipgram can be referred to as a template.
You can all of these extract relations using the --relations (-g) flag, which is to be
used in combination with --query or --querymode`. Consider the
following sample:
$ colibri-patternmodeller -i /tmp/data.colibri.patternmodel -c /tmp/hamlet.colibri.cls -q "to be" -g
Loading class decoder from file /tmp/hamlet.colibri.cls
Loading class encoder from file /tmp/hamlet.colibri.cls
Loading indexed pattern model /tmp/data.colibri.patternmodel as input model...
Post-read processing (indexedmodel)
to be 2 4 0.0117647 ngram 2 0.0769231 1:4 9:1
# PATTERN1 RELATION PATTERN2 REL.COUNT REL.FREQUENCY COUNT2
to be SUBSUMES to 2 0.5 13
to be SUBSUMES be 2 0.5 4
to be RIGHT-NEIGHBOUR-OF To {*1*} or not 1 0.25 4
to be RIGHT-NEIGHBOUR-OF To {*2*} not 1 0.25 4
to be RIGHT-NEIGHBOUR-OF not 1 0.25 5
to be RIGHT-NEIGHBOUR-OF or not 1 0.25 4
The following columns are reported, all are indented with a single tab so possible parsers can distinguish the numbers for the queried pattern itself from the relationships with other patterns.
Pattern 1 – The pattern you queried
Relation – The nature of the relationship between pattern 1 and pattern 2
Pattern 2 – The pattern that is related to the queried pattern
Relation Count – The number of times pattern 1 and pattern 2 occur in this relation
Relation Frequency – The number of times pattern 1 and pattern 2 occur in this relationas a fraction of all relations of this type
Count 2 – The absolute number of occurrences of pattern 2 in the model
Co-occurrence¶
Co-occurrence in Colibri-Core measures which patterns co-occur on the same line (i.e. usually corresponding to a sentence or whatever structural unit you decided upon when encoding your corpus). Co-occurrence is another relation in addition to the ones described in the previous section.
The degree of co-occurrence can be expressed as either an absolute occurrence
number (--cooc or -C), for a normalised mutual pointwise information
(--npmi or -Y). Both flags take a threshold, setting the threshold too
low, specially for npmi, may cause very high memory usage. The following syntax
would show all patterns that occur at least five times in the same sentence.
Note that the order of the pattern pairs does not matter; if there are two
patterns X and Y then result X Y or Y X would be the same, yet only one of them
is included in the output to prevent duplicating information:
$ colibri-patternmodeller -i /tmp/data.colibri.patternmodel -c /tmp/hamlet.colibri.cls -C 5
The colibri-cooc script provides a shortcut for this.
Finding specific patterns¶
If you have a very specific, and limited, list of patterns you want to find in large corpus data, then you can create a constraint model based on such a pattern list, and apply to your test data. This follows the same paradigm as shown in the section on training and testing.
To make a model from a pattern list, create a file with one pattern per line.
The patterns can be skipgrams or flexgrams using {*} and {**} to mark
gaps.
As always, it is vital the test and training corpus share the same classfile. You can create one in the test data first and then extend it on the pattern list:
$ colibri-classencode -o test testcorpus.txt
$ colibri-classencode -c test.colibri.cls -e -o patternlist patternlist.txt
The extended class file will be patternlist.colibri.cls. To build a model
on the patternlist, in which each line contains one pattern only, use the
--patternlist flag:
$ colibri-patternmodeller –datafile patternlist.colibri.dat –outputmodel patternlist.colibri.patternmodel –patternlist
We can now load the patternlist model as a constraint, and build on the test data:
$ colibri-patternmodeller --constrainted --inputmodel patternlist.colibri.patternmodel --datafile test.colibri.cls --classfile test.colibri.dat --outputmodel test.colibri.patternmodel
For skipgram support, add --skipgrams to this last command. If you want to
immediately print the output and see how skipgrams are instantiated, add
--instantiate --print.
The colibri-findpatterns script provides a shortcut for this procedure.
Architecture Overview¶
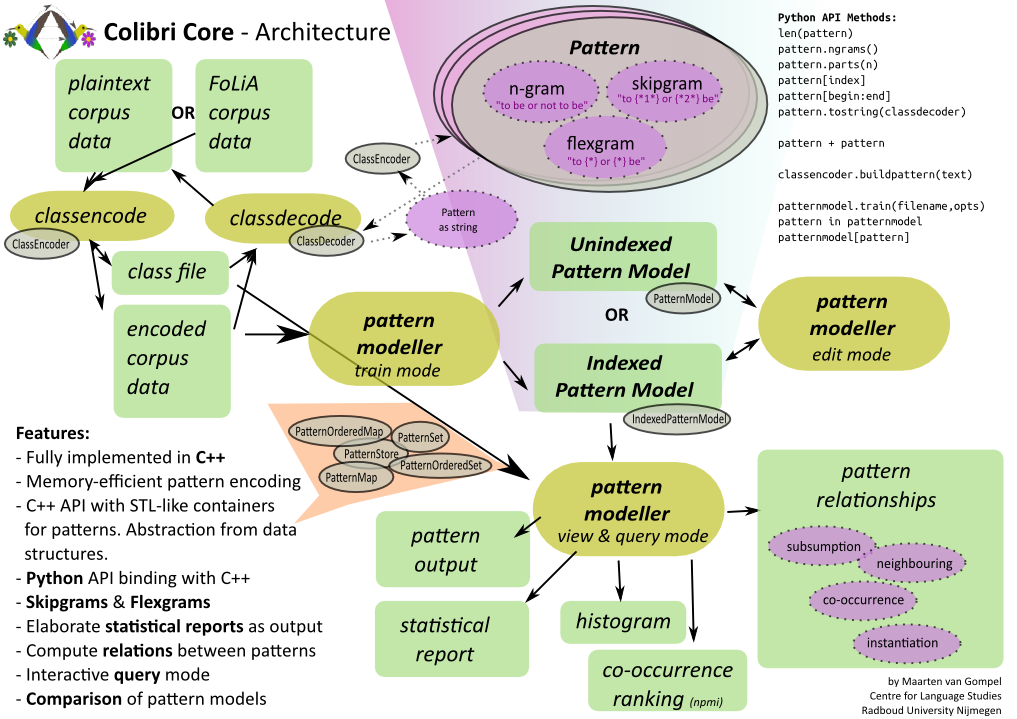
Python Tutorial¶
Colibri Core offers both a C++ API as well as a Python API. It exposes all of the functionality, and beyond, of the tools outlined above. The Python API binds with the C++ code, and although it is more limited than the C++ API, it still offers most higher-level functionality. The Colibri Core binding between C++ and Python is written in Cython.
A Python tutorial for Colibri Core is available in the form of an IPython Notebook, meaning that you can interactively run it and play with. You can go to the static, read-only, version by clicking here
Python API Reference¶
- class colibricore.AlignedPatternDict_int32¶
Bases:
objectThis is a simple low-level dictionary that takes Pattern instances as keys, and integer (unsigned 64 bit) as value. For complete pattern models, use IndexedPatternModel or UnindexPatternModel instead.
- __contains__()¶
Test if the pattern or the combination of patterns is in the aligned dictionary:
pattern in aligneddict
Or:
(pattern1,pattern2) in aligneddict
- __delitem__(key, /)¶
Delete self[key].
- __getitem__()¶
Retrieve the item, item is a two-tuple of Pattern instances.
aligneddict[(pattern1,pattern2)]
- Parameters:
item – A two tuple of Pattern instances
- __iter__()¶
Iterate over all patterns in the dictionary. If you want to iterate over pattern pairs, use pairs() instead, to iterate over the children for a specific pattern, use children()
- __len__()¶
Return the total number of patterns in the dictionary. If you want the length for a particular pattern, use childcount(pattern)
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setitem__(key, value, /)¶
Set self[key] to value.
- __setstate__()¶
- childcount()¶
Returns the number of children for the specified pattern. Use children(pattern) to iterate over them.
- children()¶
Iterate over all patterns in the dictionary. If you want to iterate over pattern pairs, use pairs() instead
- getpair()¶
- has()¶
- haspair()¶
- items()¶
Iterate over all pattern pairs and their values in the dictionary. Yields (pattern1,pattern2,value) tuples
- read()¶
- setpair()¶
- write()¶
- class colibricore.BasicPatternAlignmentModel¶
Bases:
objectPattern Alignment Model, maps patterns to pattern to score vectors (float)
Initialise an alignment model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri alignmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- __bool__()¶
True if self else False
- __contains__()¶
Tests if a pattern is in the model:
- Parameters:
pattern (Pattern or 2-tuple of patterns) – A pattern or a pair of patterns
- Return type:
bool
Example:
sourcepattern in alignmodel (sourcepattern, targetpattern) in alignmodel
- __getitem__()¶
Retrieves the value for the pattern
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int (for Unindexed Models), IndexData (for Indexed models)
Example (unindexed model):
occurrences = model[pattern]
- __init__()¶
Initialise an alignment model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri alignmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- __iter__()¶
Iterates over all source patterns in the model.
Example:
for sourcepattern in alignmodel: print(pattern.tostring(classdecoder))
- __len__()¶
Returns the total number of distinct patterns in the model
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- add()¶
Add a pattern to the unindexed model
- Parameters:
pattern (Pattern) – The source pattern to add
pattern – The target pattern to add
- items()¶
Iterate over all patterns and PatternVectors in this model, yields (Pattern, PatternVector) pairs
- load()¶
Load an alignment model from file
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri alignmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- maxlength()¶
Returns the maximum pattern length in the model
- minlength()¶
Returns the minimum pattern length in the model
- pairs()¶
Iterate over sourcepattern, targetpattern pairs
- read()¶
Alias for load
- tokens()¶
Returns the total number of tokens in the training data
- type()¶
Returns the model type (10 = UNINDEXED, 20 = INDEXED)
- types()¶
Returns the total number of distinct word types in the training data
- version()¶
Return the version of the model type
- write()¶
Write an alignment model to file
- Parameters:
filename (str) – The name of the file to write to
- class colibricore.Category¶
Bases:
objectPattern Category
- FLEXGRAM = 3¶
- NGRAM = 1¶
- SKIPGRAM = 2¶
- __dict__ = mappingproxy({'__module__': 'colibricore', '__doc__': 'Pattern Category', 'NGRAM': 1, 'SKIPGRAM': 2, 'FLEXGRAM': 3, '__dict__': <attribute '__dict__' of 'Category' objects>, '__weakref__': <attribute '__weakref__' of 'Category' objects>, '__annotations__': {}})¶
- __module__ = 'colibricore'¶
- __weakref__¶
list of weak references to the object (if defined)
- class colibricore.ClassDecoder¶
Bases:
objectThe Class Decoder allows Patterns to be decoded back to their string representation. An instance of ClassDecoder is passed to Pattern.tostring()
- __init__(*args, **kwargs)¶
- __len__()¶
Returns the total number of classes
- __new__(**kwargs)¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- decodefile()¶
- filename()¶
- class colibricore.ClassEncoder¶
Bases:
objectThe class encoder allows patterns to be built from their string representation. Load in a class file and invoke the
buildpattern()method- __init__(*args, **kwargs)¶
- __len__()¶
Returns the total number of classes
- __new__(**kwargs)¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- build()¶
Builds a class encoder from a plain-text corpus (utf-8). Equivalent to a call to processcorpus() followed by buildclasses()
- buildclasses()¶
Build classes, call after processing all corpora with processcorpus()
- buildpattern()¶
Builds a pattern: converts a string representation into a Pattern
- Parameters:
text (str) – The actual text of the pattern
allowunknown (bool) – Encode unknown classes as ‘unknown’, a single class for all, rather than failing with an exception if a word type is unseen (bool, default=False)
autoaddunknown (bool) – Automatically add unknown classes to the model (bool, default=False)
- Return type:
- encodefile()¶
Encodes the specified sourcefile according to the classer (as targetfile)
- Parameters:
sourcefile (str) – Source filename
targetfile – Target filename
allowunknown (bool) – Encode unknown classes as ‘unknown’, a single class for all, rather than failing with an exception if a word type is unseen (bool, default=False)
addunknown (bool) – Add unknown classes to the class encoder (bool, default=False)
append (bool) – Append to file (bool, default=False)
- filename()¶
- processcorpus()¶
Process a corpus, call buildclasses() when finished with all corpora
- save()¶
- class colibricore.HashOrderedIndexedPatternModel¶
Bases:
objectIndexed Pattern Model. Implemented using an ordered map
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __bool__()¶
True if self else False
- __contains__()¶
Tests if a pattern is in the model:
- Parameters:
pattern (Pattern) – A pattern
- Return type:
bool
Example:
pattern in patternmodel
- __getitem__()¶
Retrieves the value for the pattern
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int (for Unindexed Models), IndexData (for Indexed models)
Example (unindexed model):
occurrences = model[pattern]
- __init__()¶
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __iter__()¶
Iterate over all patterns in this model
- __len__()¶
Returns the total number of distinct patterns in the model
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- add()¶
Add a pattern to the indexed model :param pattern: The pattern to add :type pattern: Pattern :param indices: list (or other iterable) of 2-tuples specifying the (sentence,index) of each occurrence of the pattern
- computeflexgrams_fromcooc()¶
Compute flexgrams based on co-occurence. The threshold is expressed in normalised pointwise mutual information. Returns the number of flexgrams found. Flexgrams contain at only one gap using this method.
- computeflexgrams_fromskipgrams()¶
Compute flexgrams from skipgrams in the model. Returns the number of flexgrams found.
- coverage()¶
Returns the number of tokens all instances of the specified pattern cover in the training data, as a fraction of the total amount of tokens
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- coveragecount()¶
Returns the number of tokens all instances of the specified pattern cover in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- covered()¶
- filter()¶
Generator over patterns occurring over the set occurrence threshold (and of specified category and size if set to values above 0). This is faster than iterating and filtering manually! Will return (pattern, occurrencecount) tuples (unsorted)
- frequency()¶
Returns the frequency of the pattern within its category (ngram/skipgram/flexgram) and exact size class. For a bigram it will thus return the bigram frequency.
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- getcooc()¶
Get left-side co-occurrences for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getdata()¶
Return the indices at which the pattern occurs
- getinstance()¶
Gets a specific instance of a pattern (skipgram or flexgram), at the specified position. Raises a KeyError when not found.
- getinstances()¶
Get templates (abstracting skipgrams) for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getleftcooc()¶
Get left-side co-occurrences for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getleftneighbours()¶
Get left neighbours for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- getreverseindex()¶
Generator over all patterns occurring at the specified index reference
- Parameters:
indexreference – a (sentence, tokenoffset) tuple
- getreverseindex_bysentence()¶
Generator over all patterns occurring in the specified sentence
- Parameters:
sentence – a sentence number
- getrightcooc()¶
Get right-side co-occurrences for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getrightneighbours()¶
Get right neighbours for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- getskipcontent()¶
Get skip content for the specified pattern :param pattern: The pattern :type pattern: Pattern :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getsubchildren()¶
Get subsumption children for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- getsubparents()¶
Get subsumption parents for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- gettemplates()¶
Get templates (abstracting skipgrams) for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- histogram()¶
Generator over a histogram of occurrence count data, produces (occurrencecount, frequency) tuples. A minimum threshold may be configured, or a cap on total number of occurrences may be specified (to get only the top occurrences). The histogram can be constrained by category and/or pattern size (if set to >0 values)
- items()¶
Iterate over all patterns and their index data (IndexedData instances) in this model
- load()¶
Load a patternmodel from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- loadconstrainedbyindexedmodel()¶
- loadconstrainedbysetmodel()¶
- loadconstrainedbyunindexedmodel()¶
- loadreverseindex()¶
- maxlength()¶
Returns the maximum pattern length in the model
- minlength()¶
Returns the minimum pattern length in the model
- occurrencecount()¶
Returns the number of times the specified pattern occurs in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- outputrelations()¶
Compute and output (to stdout) all relations for the specified pattern:
- Parameters:
pattern (Pattern) – The pattern to output relations for
decoder (ClassDecoder) – The class decoder
- printhistogram()¶
Print a histogram to stdout
- printmodel()¶
Print the entire pattern model to stdout, a detailed overview
- Parameters:
decoder (ClassDecoder) – The class decoder
- prune()¶
Prune all patterns occurring below the threshold.
- Parameters:
threshold (int) – the threshold value (minimum number of occurrences)
n (int) – prune only patterns of the specified size, use 0 (default) for no size limitation
- read()¶
Alias for load
- report()¶
Print a detailed statistical report to stdout
- reverseindex()¶
Returns the reverseindex associated with the model, this will be an instance of IndexedCorpus. Use getreverseindex( (sentence, token) ) instead if you want to query the reverse index.
- tokens()¶
Returns the total number of tokens in the training data
- top()¶
Generator over the top [amount] most occurring patterns (of specified category and size if set to values above 0). This is faster than iterating manually! Will return (pattern, occurrencecount) tuples (unsorted). Note that this may return less than the specified amount of patterns if there are multiple patterns with the same occurrence count in its tail.
- totaloccurrencesingroup()¶
Returns the total number of occurrences in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total occurrence count over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalpatternsingroup()¶
Returns the total number of distinct patterns in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of distrinct skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totaltokensingroup()¶
Returns the total number of covered tokens in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered tokens over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalwordtypesingroup()¶
Returns the total number of covered word types (unigram types) in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered word types over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- train()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (None, UnindexedPatternModel, IndexedPatternModel, PatternSetModel) – A patternmodel or patternsetmodel to constrain training (default None)
- train_filtered()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
filterset – An instance of PatternSet. A limited set of skipgrams/flexgrams to use as a filter, patterns will only be included if they are an instance of a skipgram in this list (i.e. disjunctive). Ngrams can also be included as filters, if a pattern subsumes one of the ngrams in the filter, it counts as a match (or if it matches it exactly).
- trainconstrainedbyalignmodel()¶
- trainconstrainedbyindexedmodel()¶
- trainconstrainedbypatternsetmodel()¶
- trainconstrainedbyunindexedmodel()¶
- type()¶
Returns the model type (10 = UNINDEXED, 20 = INDEXED)
- types()¶
Returns the total number of distinct word types in the training data
- version()¶
Return the version of the model type
- write()¶
Write a patternmodel to file
- Parameters:
filename (str) – The name of the file to write to
- class colibricore.IndexedCorpus¶
Bases:
objectAn indexed version of a corpus, reads an entire corpus (colibri.dat file) in memory
- Parameters:
filename – The name of the colibri.dat file to load
- __contains__()¶
Test if the indexreference, a (sentence,tokenoffset) tuple is in the corpus.
- __getitem__()¶
Retrieve the token Pattern given a (sentence, tokenoffset) tuple
- __init__()¶
- Parameters:
filename – The name of the colibri.dat file to load
- __iter__()¶
Iterate over all indexes in the corpus, generator over (sentence, tokenoffset) tuples
- __len__()¶
Return the total number of tokens in the corpus
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- filename()¶
- findpattern()¶
Generator over the indexes in the corpus where this pattern is found. Note that this is much slower than using the forward index on an IndexedPatternModel!!!
- Parameters:
pattern (Pattern) – The pattern to find
sentence (int) – Set to a non-zero value to limit the search to a single sentence (default: 0, search entire corpus)
instantiate (bool) – Instantiate all skipgrams and flexgrams (i.e, return n-grams) (default: False)
- Return type:
generator over ((sentence, tokenoffset),pattern) tuples
- getinstance()¶
Gets a specific instance of a pattern (skipgram or flexgram), at the specified position. Raises a KeyError when not found.
- getsentence()¶
Get the specified sentence as a pattern, raises KeyError when the sentence, or tokens therein, does not exist
- items()¶
Iterate over all indexes and their unigram patterns. Yields ((sentence,tokenoffset), unigrampattern) tuples
- sentencecount()¶
Returns the number of sentences. ( The C++ equivalent is called sentences() )
- sentencelength()¶
Returns the length of the specified sentences, raises KeyError when it doesn’t exist
- sentences()¶
Iterates over all sentences, returning each as a pattern
- class colibricore.IndexedData¶
Bases:
objectIndexedData is essentially a set of indexes in the form of (sentence,token) tuples, sentence is generally 1-indexed, token is always 0-indexed. It is used by Indexed Pattern Models to keep track of exact occurrences of all the patterns. Use len() to if you’re merely interested in the number of occurrences, rather than their exact wherabouts.
- __bool__()¶
True if self else False
- __contains__()¶
Tests if the specified (sentence,token) tuple is in the set
- __int__()¶
int(self)
- __iter__()¶
Iterate over all (sentence,token) tuples in the set
- __len__()¶
Returns the number of occurrences, i.e. the length of the set
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- class colibricore.IndexedPatternModel¶
Bases:
objectIndexed Pattern Model. Implemented using a hash map (dictionary)
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __bool__()¶
True if self else False
- __contains__()¶
Tests if a pattern is in the model:
- Parameters:
pattern (Pattern) – A pattern
- Return type:
bool
Example:
pattern in patternmodel
- __getitem__()¶
Retrieves the value for the pattern
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int (for Unindexed Models), IndexData (for Indexed models)
Example (unindexed model):
occurrences = model[pattern]
- __init__()¶
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __iter__()¶
Iterate over all patterns in this model
- __len__()¶
Returns the total number of distinct patterns in the model
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- add()¶
Add a pattern to the indexed model :param pattern: The pattern to add :type pattern: Pattern :param indices: list (or other iterable) of 2-tuples specifying the (sentence,index) of each occurrence of the pattern
- computeflexgrams_fromcooc()¶
Compute flexgrams based on co-occurence. The threshold is expressed in normalised pointwise mutual information. Returns the number of flexgrams found. Flexgrams contain at only one gap using this method.
- computeflexgrams_fromskipgrams()¶
Compute flexgrams from skipgrams in the model. Returns the number of flexgrams found.
- coverage()¶
Returns the number of tokens all instances of the specified pattern cover in the training data, as a fraction of the total amount of tokens
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- coveragecount()¶
Returns the number of tokens all instances of the specified pattern cover in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- covered()¶
- filter()¶
Generator over patterns occurring over the set occurrence threshold (and of specified category and size if set to values above 0). This is faster than iterating and filtering manually! Will return (pattern, occurrencecount) tuples (unsorted)
- frequency()¶
Returns the frequency of the pattern within its category (ngram/skipgram/flexgram) and exact size class. For a bigram it will thus return the bigram frequency.
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- getcooc()¶
Get left-side co-occurrences for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getdata()¶
Return the indices at which the pattern occurs
- getinstance()¶
Gets a specific instance of a pattern (skipgram or flexgram), at the specified position. Raises a KeyError when not found.
- getinstances()¶
Get templates (abstracting skipgrams) for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getleftcooc()¶
Get left-side co-occurrences for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getleftneighbours()¶
Get left neighbours for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- getreverseindex()¶
Generator over all patterns occurring at the specified index reference
- Parameters:
indexreference – a (sentence, tokenoffset) tuple
- getreverseindex_bysentence()¶
Generator over all patterns occurring in the specified sentence
- Parameters:
sentence – a sentence number
- getrightcooc()¶
Get right-side co-occurrences for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getrightneighbours()¶
Get right neighbours for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- getskipcontent()¶
Get skip content for the specified pattern :param pattern: The pattern :type pattern: Pattern :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- getsubchildren()¶
Get subsumption children for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- getsubparents()¶
Get subsumption parents for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :param category: Constrain by patterns of the specified category (colibricore.Category.NGRAM,colibricore.Category.SKIPGRAM, colibricore.Category.FLEXGRAM) :param size: Constrain by patterns of the specified size :type size: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrences for this particularrelationship
- gettemplates()¶
Get templates (abstracting skipgrams) for the specified pattern :param pattern: The pattern :type pattern: Pattern :param occurrencethreshold: Constrain by patterns occurring at least this many times in this relationship (default: 0, unconstrained) :type occurrencethreshold: int :rtype: generator over (Pattern,value) tuples. The values correspond to the number of occurrence for this particularrelationship
- histogram()¶
Generator over a histogram of occurrence count data, produces (occurrencecount, frequency) tuples. A minimum threshold may be configured, or a cap on total number of occurrences may be specified (to get only the top occurrences). The histogram can be constrained by category and/or pattern size (if set to >0 values)
- items()¶
Iterate over all patterns and their index data (IndexedData instances) in this model
- load()¶
Load a patternmodel from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- loadconstrainedbyindexedmodel()¶
- loadconstrainedbysetmodel()¶
- loadconstrainedbyunindexedmodel()¶
- loadreverseindex()¶
- maxlength()¶
Returns the maximum pattern length in the model
- minlength()¶
Returns the minimum pattern length in the model
- occurrencecount()¶
Returns the number of times the specified pattern occurs in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- outputrelations()¶
Compute and output (to stdout) all relations for the specified pattern:
- Parameters:
pattern (Pattern) – The pattern to output relations for
decoder (ClassDecoder) – The class decoder
- printhistogram()¶
Print a histogram to stdout
- printmodel()¶
Print the entire pattern model to stdout, a detailed overview
- Parameters:
decoder (ClassDecoder) – The class decoder
- prune()¶
Prune all patterns occurring below the threshold.
- Parameters:
threshold (int) – the threshold value (minimum number of occurrences)
n (int) – prune only patterns of the specified size, use 0 (default) for no size limitation
- read()¶
Alias for load
- report()¶
Print a detailed statistical report to stdout
- reverseindex()¶
Returns the reverseindex associated with the model, this will be an instance of IndexedCorpus. Use getreverseindex( (sentence, token) ) instead if you want to query the reverse index.
- tokens()¶
Returns the total number of tokens in the training data
- top()¶
Generator over the top [amount] most occurring patterns (of specified category and size if set to values above 0). This is faster than iterating manually! Will return (pattern, occurrencecount) tuples (unsorted). Note that this may return less than the specified amount of patterns if there are multiple patterns with the same occurrence count in its tail.
- totaloccurrencesingroup()¶
Returns the total number of occurrences in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total occurrence count over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalpatternsingroup()¶
Returns the total number of distinct patterns in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of distrinct skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totaltokensingroup()¶
Returns the total number of covered tokens in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered tokens over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalwordtypesingroup()¶
Returns the total number of covered word types (unigram types) in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered word types over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- train()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (None, UnindexedPatternModel, IndexedPatternModel, PatternSetModel) – A patternmodel or patternsetmodel to constrain training (default None)
- train_filtered()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
filterset – An instance of PatternSet. A limited set of skipgrams/flexgrams to use as a filter, patterns will only be included if they are an instance of a skipgram in this list (i.e. disjunctive). Ngrams can also be included as filters, if a pattern subsumes one of the ngrams in the filter, it counts as a match (or if it matches it exactly).
- trainconstrainedbyalignmodel()¶
- trainconstrainedbyindexedmodel()¶
- trainconstrainedbypatternsetmodel()¶
- trainconstrainedbyunindexedmodel()¶
- type()¶
Returns the model type (10 = UNINDEXED, 20 = INDEXED)
- types()¶
Returns the total number of distinct word types in the training data
- version()¶
Return the version of the model type
- write()¶
Write a patternmodel to file
- Parameters:
filename (str) – The name of the file to write to
- class colibricore.OrderedUnindexedPatternModel¶
Bases:
objectUnindexed Pattern Model, implemented using an ordered map, less flexible and powerful than its indexed counterpart, but smaller memory footprint
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __bool__()¶
True if self else False
- __contains__()¶
Tests if a pattern is in the model:
- Parameters:
pattern (Pattern) – A pattern
- Return type:
bool
Example:
pattern in patternmodel
- __getitem__()¶
Retrieves the value for the pattern
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int (for Unindexed Models), IndexData (for Indexed models)
Example (unindexed model):
occurrences = model[pattern]
- __init__()¶
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __iter__()¶
Iterates over all patterns in the model. Also consider using the filter() or top() methods if they suit your needs, as they will be faster than doing it manually.
Example:
for pattern in model: print(pattern.tostring(classdecoder))
- __len__()¶
Returns the total number of distinct patterns in the model
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- add()¶
Add a pattern to the unindexed model
- Parameters:
pattern (Pattern) – The pattern to add
count (int) – The number of occurrences
- coverage()¶
Returns the number of tokens all instances of the specified pattern cover in the training data, as a fraction of the total amount of tokens
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- coveragecount()¶
Returns the number of tokens all instances of the specified pattern cover in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- filter()¶
Generator over patterns occurring over the set occurrence threshold (and of specified category and size if set to values above 0). This is faster than iterating and filtering manually! Will return (pattern, occurrencecount) tuples (unsorted)
- frequency()¶
Returns the frequency of the pattern within its category (ngram/skipgram/flexgram) and exact size class. For a bigram it will thus return the bigram frequency.
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- getdata()¶
- getinstance()¶
Gets a specific instance of a pattern (skipgram or flexgram), at the specified position. Raises a KeyError when not found.
- getreverseindex()¶
Generator over all patterns occurring at the specified index reference
- Parameters:
indexreference – a (sentence, tokenoffset) tuple
- getreverseindex_bysentence()¶
Generator over all patterns occurring in the specified sentence
- Parameters:
sentence – a sentence number
- histogram()¶
Generator over a histogram of occurrence count data, produces (occurrencecount, frequency) tuples. A minimum threshold may be configured, or a cap on total number of occurrences may be specified (to get only the top occurrences). The histogram can be constrained by category and/or pattern size (if set to >0 values)
- items()¶
Iterate over all patterns and their occurrence count in this model
- load()¶
Load a patternmodel from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- loadconstrainedbyindexedmodel()¶
- loadconstrainedbysetmodel()¶
- loadconstrainedbyunindexedmodel()¶
- loadreverseindex()¶
- maxlength()¶
Returns the maximum pattern length in the model
- minlength()¶
Returns the minimum pattern length in the model
- occurrencecount()¶
Returns the number of times the specified pattern occurs in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- printhistogram()¶
Print a histogram to stdout
- printmodel()¶
Print the entire pattern model to stdout, a detailed overview
- Parameters:
decoder (ClassDecoder) – The class decoder
- prune()¶
Prune all patterns occurring below the threshold.
- Parameters:
threshold (int) – the threshold value (minimum number of occurrences)
n (int) – prune only patterns of the specified size, use 0 (default) for no size limitation
- read()¶
Alias for load
- report()¶
Print a detailed statistical report to stdout
- reverseindex()¶
Returns the reverseindex associated with the model, this will be an instance of IndexedCorpus. Use getreverseindex( (sentence, token) ) instead if you want to query the reverse index.
- tokens()¶
Returns the total number of tokens in the training data
- top()¶
Generator over the top [amount] most occurring patterns (of specified category and size if set to values above 0). This is faster than iterating manually! Will return (pattern, occurrencecount) tuples (unsorted). Note that this may return less than the specified amount of patterns if there are multiple patterns with the same occurrence count in its tail.
- totaloccurrencesingroup()¶
Returns the total number of occurrences in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total occurrence count over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalpatternsingroup()¶
Returns the total number of distinct patterns in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of distrinct skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totaltokensingroup()¶
Returns the total number of covered tokens in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered tokens over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalwordtypesingroup()¶
Returns the total number of covered word types (unigram types) in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered word types over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- train()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (None, UnindexedPatternModel, IndexedPatternModel, PatternSetModel) – A patternmodel or patternsetmodel to constrain training (default None)
- train_filtered()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
filterset – An instance of PatternSet. A limited set of skipgrams/flexgrams to use as a filter, patterns will only be included if they are an instance of a skipgram in this list (i.e. disjunctive). Ngrams can also be included as filters, if a pattern subsumes one of the ngrams in the filter, it counts as a match (or if it matches it exactly).
- trainconstrainedbyalignmodel()¶
- trainconstrainedbyindexedmodel()¶
- trainconstrainedbypatternsetmodel()¶
- trainconstrainedbyunindexedmodel()¶
- type()¶
Returns the model type (10 = UNINDEXED, 20 = INDEXED)
- types()¶
Returns the total number of distinct word types in the training data
- version()¶
Return the version of the model type
- write()¶
Write a patternmodel to file
- Parameters:
filename (str) – The name of the file to write to
- class colibricore.Pattern¶
Bases:
objectThe Pattern class contains an ngram, skipgram or flexgram, and allows a wide variety of actions to be performed on it. It is stored in a memory-efficient fashion and facilitating fast operation and comparison. Use ClassEncoder.buildpattern to build a pattern.
- __add__()¶
Concatenate two patterns together, forming a larger one:
pattern3 = pattern1 + pattern2
- __bool__()¶
True if self else False
- __bytes__()¶
- __contains__()¶
Check if the specified pattern occurs within this larger pattern.
- Parameters:
pattern (Pattern) – the subpattern to look for
- Return type:
bool
Example:
subpattern in pattern
- __copy__()¶
Produces a copy of the pattern (deep):
pattern2 = copy(pattern)
- __deepcopy__()¶
Produces a copy of the pattern (deep):
pattern2 = copy(pattern)
- __eq__(value, /)¶
Return self==value.
- __ge__(value, /)¶
Return self>=value.
- __getitem__()¶
Retrieves a word/token from the pattern:
unigram = pattern[index]
Or retrieves a subpattern:
subpattern = pattern[begin:end]
- Parameters:
item – an index or slice
- Return type:
a Pattern instance
- __getstate__()¶
- __gt__(value, /)¶
Return self>value.
- __hash__()¶
Returns the hashed value for this pattern
- __iter__()¶
Iterates over all words/tokens in this pattern
- __le__(value, /)¶
Return self<=value.
- __len__()¶
Returns the length of the pattern in words/tokens:
len(pattern)
- __lt__(value, /)¶
Return self<value.
- __ne__(value, /)¶
Return self!=value.
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __radd__(value, /)¶
Return value+self.
- __reduce_cython__()¶
- __setstate__()¶
- __setstate_cython__()¶
- bindboundary()¶
- bindflex()¶
- bindskip()¶
- bindunk()¶
- bytesize()¶
Returns the number of bytes used to encode this pattern in memory
- category()¶
Returns the category of this pattern :rtype: Category.NGRAM (1), Category.SKIPGRAM (2), or Category.FLEXGRAM (3)
- concat()¶
- gaps()¶
Generator iterating over the gaps in a skipgram or flexgram, return a tuple (begin,length) for each. For flexgrams, the minimum length (1) is always returned.
- Return type:
generator over (begin, length) tuples
- instanceof()¶
Is this an instantiation of the skipgram/flexgram? Instantiation is not necessarily full, aka: A ? B C is also an instantiation of A ? ? C
- isflexgram()¶
- isgap()¶
- isskipgram()¶
- ngrams()¶
Generator iterating over all ngrams of a particular size (or range thereof) that are enclosed within this pattern. Despite the name, this may also return skipgrams!
- Parameters:
n (int) – The desired size to obtain, if unspecified (0), will extract all ngrams
maxn (int) – If larger than n, will extract ngrams in the specified n range
- Return type:
generator over Pattern instances
- parts()¶
Generating iterating over the consecutive non-gappy parts in a skipgram of flexgram
- Return type:
generator over Pattern instances
- reverse()¶
Reverse the pattern (all the words will be in reverse order)
- skipcount()¶
Returns the number of gaps in this pattern
- subngrams()¶
Generator iterating over all ngrams of all sizes that are enclosed within this pattern. Despite the name, this may also return skipgrams! :param minn: minimum length (default 1) :type minn: int :param maxn: maximum length (default unlimited) :type maxn: int :rtype: generator over Pattern instances
- tolist()¶
Returns a list representing the raw classes in the pattern
- tostring()¶
Convert a Pattern back to a str
- Parameters:
decoder (ClassDecoder) – the class decoder to use
- Return type:
str
- unknown()¶
- class colibricore.PatternAlignmentModel_float¶
Bases:
objectPattern Alignment Model, maps patterns to pattern to score vectors (float)
Initialise an alignment model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri alignmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- __bool__()¶
True if self else False
- __contains__()¶
Tests if a pattern is in the model:
- Parameters:
pattern (Pattern or 2-tuple of patterns) – A pattern or a pair of patterns
- Return type:
bool
Example:
sourcepattern in alignmodel (sourcepattern, targetpattern) in alignmodel
- __getitem__()¶
Retrieves the value for the pattern
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int (for Unindexed Models), IndexData (for Indexed models)
Example (unindexed model):
occurrences = model[pattern]
- __init__()¶
Initialise an alignment model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri alignmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- __iter__()¶
Iterates over all source patterns in the model.
Example:
for sourcepattern in alignmodel: print(pattern.tostring(classdecoder))
- __len__()¶
Returns the total number of distinct patterns in the model
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- add()¶
Add a pattern to the unindexed model
- Parameters:
pattern (Pattern) – The source pattern to add
pattern – The target pattern to add
l (tuple (of floats)) – Feature tuple (homogenously typed)
- items()¶
Iterate over all patterns and PatternFeatureVectorMaps in this model
- load()¶
Load an alignment model from file
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri alignmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- maxlength()¶
Returns the maximum pattern length in the model
- minlength()¶
Returns the minimum pattern length in the model
- read()¶
Alias for load
- tokens()¶
Returns the total number of tokens in the training data
- triples()¶
Iterate over sourcepattern, targetpattern, feature triples
- type()¶
Returns the model type (10 = UNINDEXED, 20 = INDEXED)
- types()¶
Returns the total number of distinct word types in the training data
- version()¶
Return the version of the model type
- write()¶
Write an alignment model to file
- Parameters:
filename (str) – The name of the file to write to
- class colibricore.PatternDict_float¶
Bases:
objectThis is a simple low-level dictionary that takes Pattern instances as keys, and float (double) as value. For complete pattern models, use IndexedPatternModel or UnindexPatternModel instead.
- __bool__()¶
True if self else False
- __contains__()¶
Test if the pattern is in the dictionary
- __delitem__(key, /)¶
Delete self[key].
- __getitem__()¶
Retrieve the value for a pattern in the dictionary
- Parameters:
pattern (Pattern) – A pattern
- __iter__()¶
Iterate over all patterns in the dictionary
- __len__()¶
Return the total number of patterns in the dictionary
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setitem__()¶
Set the value for a pattern in the dictionary
- Parameters:
pattern – the pattern
value – its value
- __setstate__()¶
- has()¶
- items()¶
Iterate over all patterns and their values in the dictionary
- read()¶
- write()¶
- class colibricore.PatternDict_int¶
Bases:
objectThis is a simple low-level dictionary that takes Pattern instances as keys, and integer (unsigned 64 bit) as value. For complete pattern models, use IndexedPatternModel or UnindexPatternModel instead.
- __bool__()¶
True if self else False
- __contains__()¶
Test if the pattern is in the dictionary
- __delitem__(key, /)¶
Delete self[key].
- __getitem__()¶
Retrieve the value for a pattern in the dictionary
- Parameters:
pattern (Pattern) – A pattern
- __iter__()¶
Iterate over all patterns in the dictionary
- __len__()¶
Return the total number of patterns in the dictionary
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setitem__()¶
Set the value for a pattern in the dictionary
- Parameters:
pattern – the pattern
value – its value
- __setstate__()¶
- has()¶
- items()¶
Iterate over all patterns and their values in the dictionary
- read()¶
- write()¶
- class colibricore.PatternDict_int32¶
Bases:
objectThis is a simple low-level dictionary that takes Pattern instances as keys, and integer (max 32 bit, unsigned) as value. For complete pattern models, use IndexedPatternModel or UnindexPatternModel instead.
- __bool__()¶
True if self else False
- __contains__()¶
Test if the pattern is in the dictionary
- __delitem__(key, /)¶
Delete self[key].
- __getitem__()¶
Retrieve the value for a pattern in the dictionary
- Parameters:
pattern (Pattern) – A pattern
- __iter__()¶
Iterate over all patterns in the dictionary
- __len__()¶
Return the total number of patterns in the dictionary
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setitem__()¶
Set the value for a pattern in the dictionary
- Parameters:
pattern – the pattern
value – its value
- __setstate__()¶
- has()¶
- items()¶
Iterate over all patterns and their values in the dictionary
- read()¶
- write()¶
- class colibricore.PatternFeatureVectorMap_float¶
Bases:
object- __bool__()¶
True if self else False
- __contains__(key, /)¶
Return key in self.
- __delitem__(key, /)¶
Delete self[key].
- __getitem__(key, /)¶
Return self[key].
- __iter__()¶
Implement iter(self).
- __len__()¶
Return len(self).
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setitem__(key, value, /)¶
Set self[key] to value.
- __setstate__()¶
- items()¶
- setdata()¶
- class colibricore.PatternModelOptions¶
Bases:
objectOptions for Pattern model, you can get and set the following attributes:
MINTOKENS - The token threshold, patterns with an occurrence below this will be pruned
MAXLENGTH - Maximum pattern length
DOSKIPGRAMS - Compute skipgrams?
DOSKIPGRAMS_EXHAUSTIVE - Compute skipgrams exhaustively?
MINSKIPTYPES - Minimum amount of different skip content types
MAXSKIPS - The maximum amount of skips in a skipgram
DOREVERSEINDEX - Build reverse index? (default: True)
DOPATTERNPERLINE - Assume each line holds one single pattern.
MINTOKENS_UNIGRAMS - Word occurrence threshold (secondary threshold): only count patterns in which the words/unigrams occur at least this many times, only effective when the primary
MINTOKENS_SKIPGRAMS - The occurrence threshold for skipgrams, minimum amount of occurrences for a pattern to be included in a model. Defaults to the same value as MINTOKENS. Only used if DOSKIPGRAMS or DO_SKIPGRAMS_EXHAUSTIVE is set to true
DOREMOVENGRAMS - Remove n-grams from the model
DOREMOVESKIPGRAMS - Remove skipgrams from the model
DOREMOVEFLEXGRAMS - Remove flexgrams from the model
DORESET - Reset all counts before training
PRUNENONSUBSUMED - Prune all n-grams up to this length that are NOT subsumed by higher-order ngrams
PRUNESUBSUMED - Prune all n-grams up to this length that are subsumed by higher-order ngrams
DEBUG
QUIET (default: False)
These can also be passed at keyword arguments to the constructor, in a case insensitive fashion:
options = PatternModelOptions(mintokens=3)
- __delattr__(name, /)¶
Implement delattr(self, name).
- __getattr__()¶
- __getattribute__(name, /)¶
Return getattr(self, name).
- __init__(*args, **kwargs)¶
- __new__(**kwargs)¶
- __reduce__()¶
Helper for pickle.
- __setattr__(name, value, /)¶
Implement setattr(self, name, value).
- __setstate__()¶
- class colibricore.PatternSet¶
Bases:
objectThis is a simple low-level set that contains Pattern instances
- __bool__()¶
True if self else False
- __contains__()¶
Test if the pattern is in the dictionary
- Parameters:
pattern (Pattern) – A pattern
- Return type:
bool
- __iter__()¶
Iterate over all patterns in the dictionary
- __len__()¶
Return the total number of patterns in the dictionary
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- has()¶
- class colibricore.PatternSetModel¶
Bases:
objectInitialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- __bool__()¶
True if self else False
- __contains__()¶
Test if the pattern is in the dictionary
- Parameters:
pattern (Pattern) – A pattern
- Return type:
bool
- __init__()¶
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- __iter__()¶
Iterate over all patterns in the dictionary
- __len__()¶
Return the total number of patterns in the dictionary
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- has()¶
- load()¶
Load a patternmodel from file
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- read()¶
Alias for load
- write()¶
Write a patternmodel to file
- Parameters:
filename (str) – The name of the file to write to
- class colibricore.PatternVector¶
Bases:
object- __bool__()¶
True if self else False
- __contains__(key, /)¶
Return key in self.
- __iter__()¶
Implement iter(self).
- __len__()¶
Return len(self).
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- class colibricore.SmallPatternDict_int32¶
Bases:
objectThis is a simple low-level dictionary that takes Pattern instances as keys, and integer (max 32 bit, unsigned) as value. For complete pattern models, use IndexedPatternModel or UnindexPatternModel instead. This is a Small version taht allows at most 65536 patterns.
- __bool__()¶
True if self else False
- __contains__()¶
Test if the pattern is in the dictionary
- __delitem__(key, /)¶
Delete self[key].
- __getitem__()¶
Retrieve the value for a pattern in the dictionary
- Parameters:
pattern (Pattern) – A pattern
- __iter__()¶
Iterate over all patterns in the dictionary
- __len__()¶
Return the total number of patterns in the dictionary
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setitem__()¶
Set the value for a pattern in the dictionary
- Parameters:
pattern – the pattern
value – its value
- __setstate__()¶
- has()¶
- items()¶
Iterate over all patterns and their values in the dictionary
- read()¶
- write()¶
- class colibricore.TinyPatternDict_int32¶
Bases:
objectThis is a simple low-level dictionary that takes Pattern instances as keys, and integer (max 32 bit, unsigned) as value. For complete pattern models, use IndexedPatternModel or UnindexPatternModel instead. This is a tiny version that allow only up to 256 patterns.
- __bool__()¶
True if self else False
- __contains__()¶
Test if the pattern is in the dictionary
- __delitem__(key, /)¶
Delete self[key].
- __getitem__()¶
Retrieve the value for a pattern in the dictionary
- Parameters:
pattern (Pattern) – A pattern
- __iter__()¶
Iterate over all patterns in the dictionary
- __len__()¶
Return the total number of patterns in the dictionary
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setitem__()¶
Set the value for a pattern in the dictionary
- Parameters:
pattern – the pattern
value – its value
- __setstate__()¶
- has()¶
- items()¶
Iterate over all patterns and their values in the dictionary
- read()¶
- write()¶
- class colibricore.UnindexedPatternModel¶
Bases:
objectUnindexed Pattern Model, less flexible and powerful than its indexed counterpart, but smaller memory footprint
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __bool__()¶
True if self else False
- __contains__()¶
Tests if a pattern is in the model:
- Parameters:
pattern (Pattern) – A pattern
- Return type:
bool
Example:
pattern in patternmodel
- __getitem__()¶
Retrieves the value for the pattern
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int (for Unindexed Models), IndexData (for Indexed models)
Example (unindexed model):
occurrences = model[pattern]
- __init__()¶
Initialise a pattern model. Either an empty one or loading from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (IndexedPatternModel, UnindexedPatternModel or None (default)) – A model to be used as a constraint, only patterns occuring in this constraint model will be loaded/trained
reverseindex (IndexedCorpus or None) – Corpus data to use as reverse index. On indexed models, this is required for various operations, including computation of skipgrams
- __iter__()¶
Iterates over all patterns in the model. Also consider using the filter() or top() methods if they suit your needs, as they will be faster than doing it manually.
Example:
for pattern in model: print(pattern.tostring(classdecoder))
- __len__()¶
Returns the total number of distinct patterns in the model
- __new__(**kwargs)¶
- __pyx_vtable__ = <capsule object NULL>¶
- __reduce__()¶
Helper for pickle.
- __setstate__()¶
- add()¶
Add a pattern to the unindexed model
- Parameters:
pattern (Pattern) – The pattern to add
count (int) – The number of occurrences
- coverage()¶
Returns the number of tokens all instances of the specified pattern cover in the training data, as a fraction of the total amount of tokens
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- coveragecount()¶
Returns the number of tokens all instances of the specified pattern cover in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- filter()¶
Generator over patterns occurring over the set occurrence threshold (and of specified category and size if set to values above 0). This is faster than iterating and filtering manually! Will return (pattern, occurrencecount) tuples (unsorted)
- frequency()¶
Returns the frequency of the pattern within its category (ngram/skipgram/flexgram) and exact size class. For a bigram it will thus return the bigram frequency.
- Parameters:
pattern (Pattern) – A pattern
- Return type:
float
- getdata()¶
- getinstance()¶
Gets a specific instance of a pattern (skipgram or flexgram), at the specified position. Raises a KeyError when not found.
- getreverseindex()¶
Generator over all patterns occurring at the specified index reference
- Parameters:
indexreference – a (sentence, tokenoffset) tuple
- getreverseindex_bysentence()¶
Generator over all patterns occurring in the specified sentence
- Parameters:
sentence – a sentence number
- histogram()¶
Generator over a histogram of occurrence count data, produces (occurrencecount, frequency) tuples. A minimum threshold may be configured, or a cap on total number of occurrences may be specified (to get only the top occurrences). The histogram can be constrained by category and/or pattern size (if set to >0 values)
- items()¶
Iterate over all patterns and their occurrence count in this model
- load()¶
Load a patternmodel from file.
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri patternmodel file
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
- loadconstrainedbyindexedmodel()¶
- loadconstrainedbysetmodel()¶
- loadconstrainedbyunindexedmodel()¶
- loadreverseindex()¶
- maxlength()¶
Returns the maximum pattern length in the model
- minlength()¶
Returns the minimum pattern length in the model
- occurrencecount()¶
Returns the number of times the specified pattern occurs in the training data
- Parameters:
pattern (Pattern) – A pattern
- Return type:
int
- printhistogram()¶
Print a histogram to stdout
- printmodel()¶
Print the entire pattern model to stdout, a detailed overview
- Parameters:
decoder (ClassDecoder) – The class decoder
- prune()¶
Prune all patterns occurring below the threshold.
- Parameters:
threshold (int) – the threshold value (minimum number of occurrences)
n (int) – prune only patterns of the specified size, use 0 (default) for no size limitation
- read()¶
Alias for load
- report()¶
Print a detailed statistical report to stdout
- reverseindex()¶
Returns the reverseindex associated with the model, this will be an instance of IndexedCorpus. Use getreverseindex( (sentence, token) ) instead if you want to query the reverse index.
- tokens()¶
Returns the total number of tokens in the training data
- top()¶
Generator over the top [amount] most occurring patterns (of specified category and size if set to values above 0). This is faster than iterating manually! Will return (pattern, occurrencecount) tuples (unsorted). Note that this may return less than the specified amount of patterns if there are multiple patterns with the same occurrence count in its tail.
- totaloccurrencesingroup()¶
Returns the total number of occurrences in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total occurrence count over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalpatternsingroup()¶
Returns the total number of distinct patterns in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of distrinct skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totaltokensingroup()¶
Returns the total number of covered tokens in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered tokens over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- totalwordtypesingroup()¶
Returns the total number of covered word types (unigram types) in the specified group, within the specified category and/or size class, you can set either to zero (default) to consider all. Example, category=Category.SKIPGRAM and n=0 would consider give the total number of covered word types over all skipgrams.
- Parameters:
category (int) – The category constraint (Category.NGRAM, Category.SKIPGRAM, Category.FLEXGRAM or 0 for no-constraint, default)
n (int) – The size constraint (0= no constraint, default)
- Return type:
int
- train()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
constrainmodel (None, UnindexedPatternModel, IndexedPatternModel, PatternSetModel) – A patternmodel or patternsetmodel to constrain training (default None)
- train_filtered()¶
Train the patternmodel on the specified corpus data (a *.colibri.dat file)
- Parameters:
filename (str) – The name of the file to load, must be a valid colibri.dat file. Can be set to an empty string if a corpus was pre-loaded already.
options (PatternModelOptions) – An instance of PatternModelOptions, containing the options used for loading
filterset – An instance of PatternSet. A limited set of skipgrams/flexgrams to use as a filter, patterns will only be included if they are an instance of a skipgram in this list (i.e. disjunctive). Ngrams can also be included as filters, if a pattern subsumes one of the ngrams in the filter, it counts as a match (or if it matches it exactly).
- trainconstrainedbyalignmodel()¶
- trainconstrainedbyindexedmodel()¶
- trainconstrainedbypatternsetmodel()¶
- trainconstrainedbyunindexedmodel()¶
- type()¶
Returns the model type (10 = UNINDEXED, 20 = INDEXED)
- types()¶
Returns the total number of distinct word types in the training data
- version()¶
Return the version of the model type
- write()¶
Write a patternmodel to file
- Parameters:
filename (str) – The name of the file to write to
- colibricore.encode()¶
- colibricore.patternfromfile()¶
Builds a single pattern from corpus data, will ignore any newlines. You may want to use IndexedCorpus instead.
- colibricore.write()¶
Write a patternmodel to file
- Parameters:
filename (str) – The name of the file to write to